Cluefish: mining the dark matter of transcriptional data series with over-representation analysis enhanced by aggregated biological prior knowledge
Important links
Abstract
Interpreting transcriptomic data presents significant challenges, particularly in non-targeted approaches. While modern functional enrichment methods are well-suited for experimental designs involving two conditions, they are less applicable to data series. In this context, we developed Cluefish, a free and open-source, semi-automated R workflow designed for untargeted, comprehensive biological interpretation of transcriptomic data series. Cluefish applies over-representation analysis on pre-clustered protein–protein interaction networks, using clusters as anchors to identify smaller, more specific biological functions. Innovative features, including cluster merging and recovery of isolated genes through shared biological contexts, enable a more complete exploration of the data. We applied Cluefish to an in-house dataset with zebrafish exposed to a dose-gradient of dibutyl phthalate and to two published toxicology datasets featuring different organisms. Combined with DRomics, a tool for dose–response analysis—Cluefish identified gene clusters deregulated at low doses and linked to biological functions overlooked by the standard approach. Notably, it revealed that retinoid signaling disruption may be the most sensitive pathway affected by dibutyl phthalate during zebrafish development, potentially leading to morphological changes. The Cluefish workflow aims to provide valuable clues for biological hypothesis generation and experimental validation. It is freely available at https://github.com/ellfran-7/cluefish.
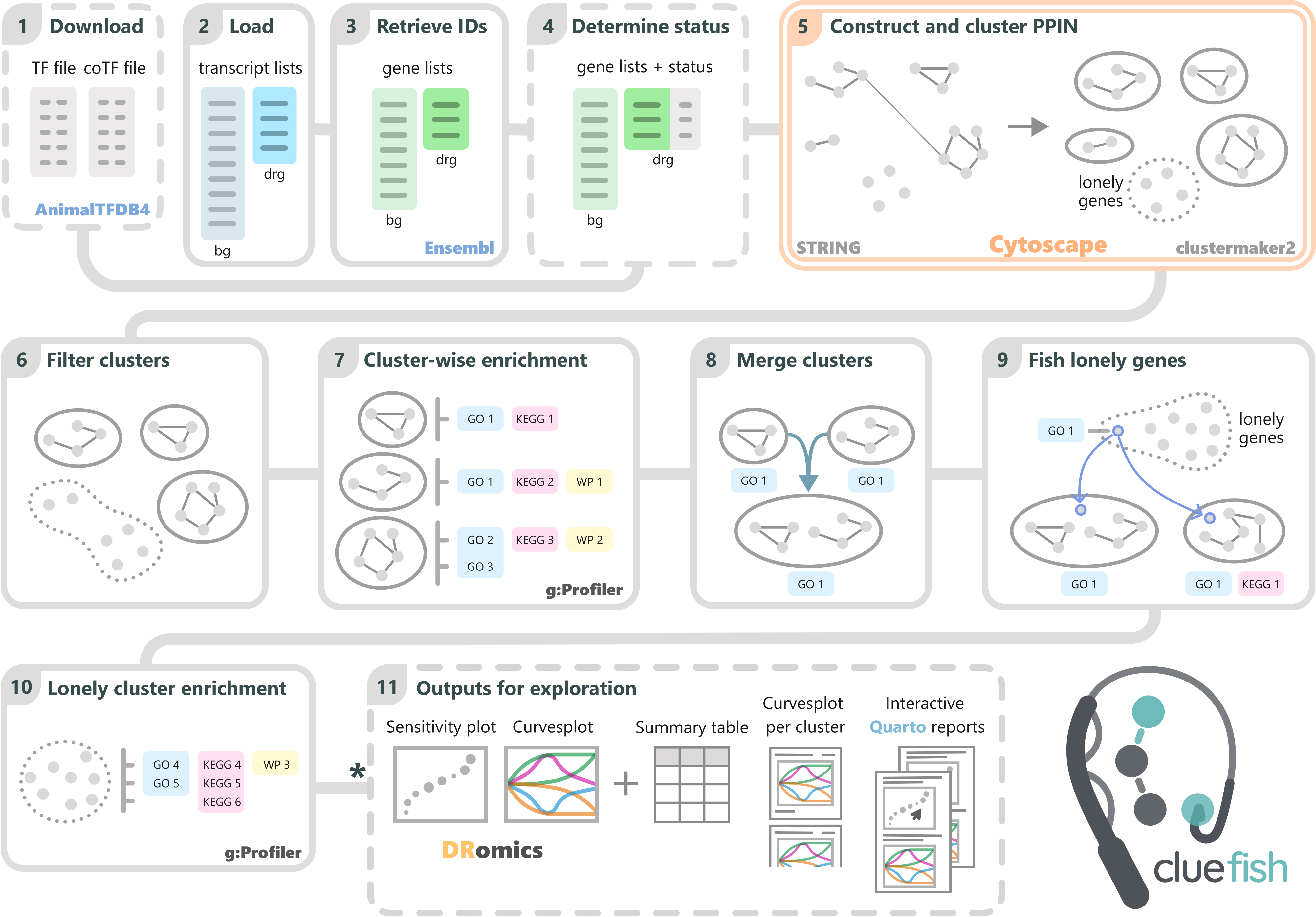
The workflow
Citation
@article{10.1093/nargab/lqaf103,
author = {Franklin, Ellis and Billoir, Elise and Veber, Philippe and Ohanessian, Jérémie and Delignette-Muller, Marie Laure and Prud’homme, Sophie Martine},
title = {Cluefish: mining the dark matter of transcriptional data series with over-representation analysis enhanced by aggregated biological prior knowledge},
journal = {NAR Genomics and Bioinformatics},
volume = {7},
number = {3},
pages = {lqaf103},
year = {2025},
month = {07},
abstract = {Interpreting transcriptomic data presents significant challenges, particularly in non-targeted approaches. While modern functional enrichment methods are well-suited for experimental designs involving two conditions, they are less applicable to data series. In this context, we developed Cluefish, a free and open-source, semi-automated R workflow designed for untargeted, comprehensive biological interpretation of transcriptomic data series. Cluefish applies over-representation analysis on pre-clustered protein–protein interaction networks, using clusters as anchors to identify smaller, more specific biological functions. Innovative features, including cluster merging and recovery of isolated genes through shared biological contexts, enable a more complete exploration of the data. We applied Cluefish to an in-house dataset with zebrafish exposed to a dose-gradient of dibutyl phthalate and to two published toxicology datasets featuring different organisms. Combined with DRomics, a tool for dose–response analysis—Cluefish identified gene clusters deregulated at low doses and linked to biological functions overlooked by the standard approach. Notably, it revealed that retinoid signaling disruption may be the most sensitive pathway affected by dibutyl phthalate during zebrafish development, potentially leading to morphological changes. The Cluefish workflow aims to provide valuable clues for biological hypothesis generation and experimental validation. It is freely available at https://github.com/ellfran-7/cluefish.},
issn = {2631-9268},
doi = {10.1093/nargab/lqaf103},
url = {https://doi.org/10.1093/nargab/lqaf103},
eprint = {https://academic.oup.com/nargab/article-pdf/7/3/lqaf103/63893022/lqaf103.pdf},
}